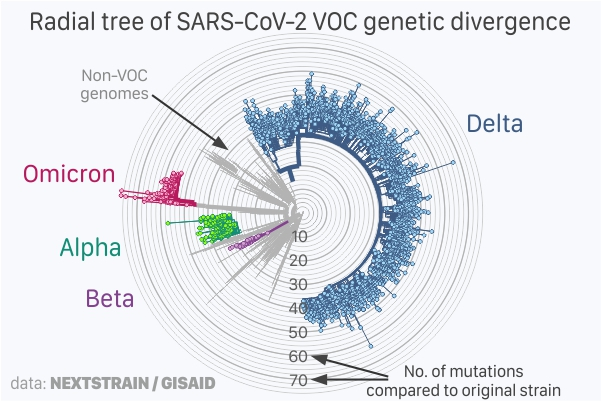
 Radial view of number of mutations in SARS-CoV-2 VOC genome sequences submitted at Gisaid, adapted from Nextstrain.;
Radial view of number of mutations in SARS-CoV-2 VOC genome sequences submitted at Gisaid, adapted from Nextstrain.;
The SARS-CoV-2 variant B.1.1.529 was first reported to the World Health Organization (WHO) from South Africa on 24 November 2021 and two days later was named “Variant of Concern (VOC) Omicron”. Belgium reported the first case in Europe on 26 November 2021 (sample collected on 22 November 2021).
On 4 January 2022, over two million new SARS-CoV-2 positive cases were reported globally, compared to fewer than 700,000 cases on 1 December 2021 – an exceptional rise compared to previous daily records. Several countries reported record-breaking daily cases in last few days.
Omicron genome (RNA sequence) has around 60 mutations compared to the original sequence (see figure above). The radial tree shows 3,449 SARS-CoV-2 genomes (December 2019 - December 2021) scaled by its genetic divergence (number of mutations) compared to the original strain. The inner circles show fewer mutations and the outer circles show a higher number of mutations, with the centre of the radial tree being the original sequence (0 mutations) and all other genomes are shown in relation to it. The tree was derived from Nextstrain on 5 December 2022.
In the case of Omicron, the spike protein alone has more than 30 mutations. These mutations are of importance and interest as they help interact and infect human cells as well as the primary target for the current generations of vaccines. Hence, the more mutations there are in the spike protein, the higher the chances that current generation of vaccine-induced immunity (antibodies) would not recognise the evolved Omicron spike protein.
Several of these mutations in the spike protein are also found in the Alpha, Beta, Gamma or Delta VOCs, with functional attributes documented as increased transmissibility, higher binding affinity and higher antibody escape.
At this point, one of the first questions that comes to mind is: How did the Omicron VOC come into being and how likely was such an eventuality, present and future?
There is no exact method or surety to find an accurate solution to these questions: however, we can start with the location and associated characteristics from where the first cases were documented. In saying so, South Africa had a low vaccination rate (less than 25% fully vaccinated in November 2021) and an estimated 20% of its population lives with AIDS (with low immunity). Low vaccination rates would mean that the current strains would continue to infect unvaccinated population and people with low immunity or immunocompromised individuals on medications would allow the virus to infect and stay longer in the body. Together, this may increase the risk of evolved strain with more mutations.
One additional factor which could have played an augmented role is the antiviral drug Molnupiravir, which was started during the Phase II/III clinical trials from October 2020 to treat mild-to-moderate COVID-19 infected adults in 23 countries, including South Africa. The participants of the clinical trials were unvaccinated and included individuals with chronic obstructive pulmonary disease or active cancer, which are also low-immunity individuals.
Molnupiravir is an oral prodrug with antiviral activity. Inside the body, Molnupiravir is metabolised and recognised by the RNA-dependent RNA polymerase (nsp12), a protein that replicates the SARS-CoV-2 genome but introduces random mutations. An active accumulation of mutations in the viral genome would mean that the translated proteins would be very different or not formed at all and thus the mutated viruses are unviable.
In nature, random mutations happen as well and this is how the other VOCs have evolved. However, slow evolutions mean only few mutations are accepted the viral genome over time. With Molnupiravir, the process is accelerated (as it randomly introduces mutations) and, in the case of immunocompromised individuals with low immunity but who are on therapy, would mean that the natural defences of the human body are low while the evolved viruses can continue to multiply.
The Food and Drug Administration (FDA) briefing document on Molnupiravir, from 30 November 2021, shows that of the 775 randomised clinical trial participants, 42.1% had contracted the Delta VOC. In other words, the mutations already present in Delta may be seen as the baseline for further mutations post treatment.
Moreover, the non-hospitalised participants for whom the nasopharyngeal (NP) samples were collected showed 0 - 46 mutations per 10,000 nucleotides sequenced on day 5 of treatment (200mg Molnupiravir capsules taken orally every twelve hours for five days, 20 capsules total) compared to 0 - 30 mutations per 10,000 nucleotides sequenced on day 5 of placebo. A total of 81 emergent spike-protein mutations were detected among 38 Molnupiravir-treated participants and nine placebo-treated participants.
The FDA report concludes that there are modest but significantly higher nucleotide mutation rates in SARS-CoV-2 genome post Molnupiravir treatment.
In summary, it remains a rare but likely scenario of a combination of all the different factors for an increased risk for an evolved strain, like Omicron, which has mutations throughout its genomic sequence.
It may therefore be prudent to revaluate continued use of Molnupiravir and similar prodrugs with antiviral activities based on RNA-dependent RNA-replication mutagenesis pathways.
The Molnupiravir clinical trial details are available at https://clinicaltrials.gov/ct2/show/NCT04575597 and the full FDA report on Molnupiravir at https://www.fda.gov/media/154418/download









